CDC Incremental Data Loading with SQL Server, Apache Iceberg and Apache Spark
Learn how to create incremental an Apache Spark setup that polls data from SQL Server its Change Data Capture (CDC) feature and saves it to Apache Iceberg
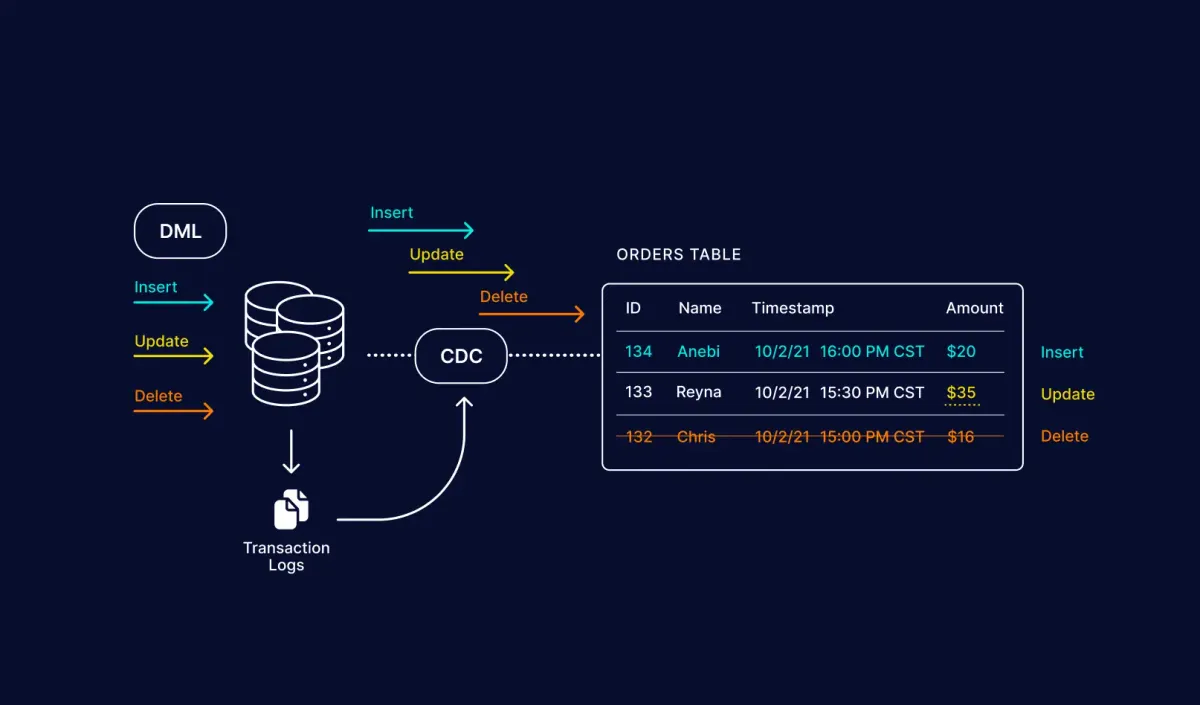
Imagine, you are working with Big Data and you just learned that the Medallion architecture exists and you start creating your bronze layer. Now suddenly, out of nowhere you are sinking your entire database each time over and over again making your pipelines run for hours instead of the usual minutes. Now you wonder: "There must be a more efficient way to handle incremental data loads right? "
What appeared to initially be presented as a walk in the park, turned out to be a battle between giants, a battle between ecosystem, a battle between people, all resulting in picking something that just works from a given viewpoint
💡 I would dare to say that this also formalizes "Architecture", where we are responsible of selecting components through the lens of Strategic viewpoint, helping you decide what's best for the business. For example, in my case, I tend to select components with focus on separation of concerns and lock-in avoidance.
Let's dive more into this and how we can find a future proof, reliable and trusted solution for this that can handle our Big Data needs at scale. But before we get started, let's first go over the different components and why I picked those.
Creating our components (Tech Stack )
SQL Server
The first component we need is a SQL Server Database that will hold our data. Besides just holding our data, it should also support a feature named "Change Data Capture" (CDC) that allows us to track changes made to the database and receive those. Something we rely heavily on for this!
Apache Iceberg
Since we want to perform large scale analytics on all our data (both structured and unstructured) we require a storage mechanism that can efficiently work with Flat Files. I currently chose here for Apache Iceberg, as it is created for large scale analytics on "Lakehouse" and Data Streaming and integrates with all the major platforms.
Many might wonder, why Iceberg and not Delta? Well Delta is great as well, but was created with a more commercial oriented approach, and more importantly Iceberg is great at schema evolution and partitioning. Besides this, it seems the community is also more keen on Iceberg, which is seeing a tremendous increase in users over time.
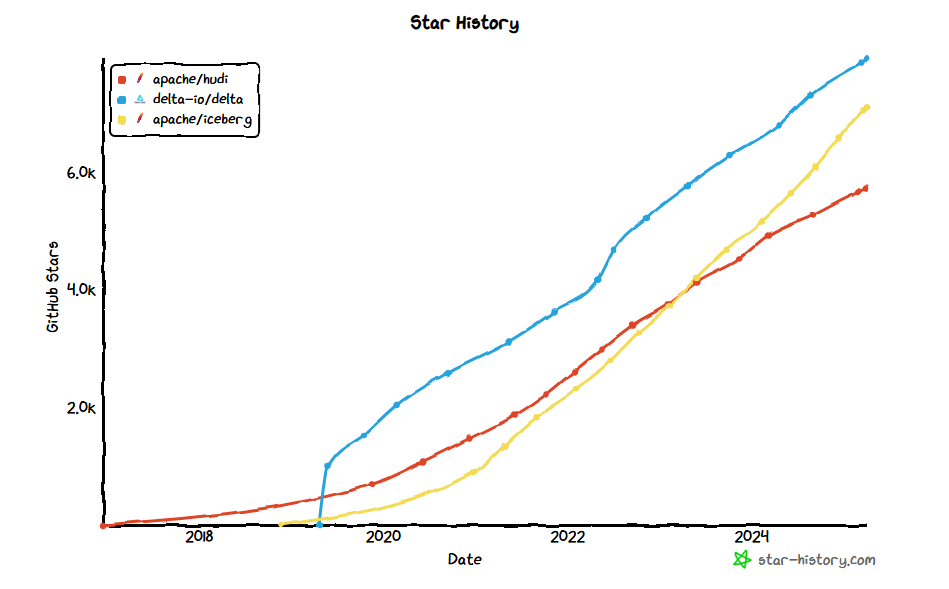
This doesn't mean that Delta is bad! But currently, seeing the current state of the ecosystem, I would like to avoid a lock-in or draw-in into a given ecosystem. This is also reflected by the larger organizations such as Apple, Microsoft, AWS and others adopting Iceberg.
💡 Databricks acquired "Tabular" recently, which was the core maintainer of Apache Iceberg (smart move). So the current future of both Delta and Iceberg is uncertain and time will tell what will happen here.
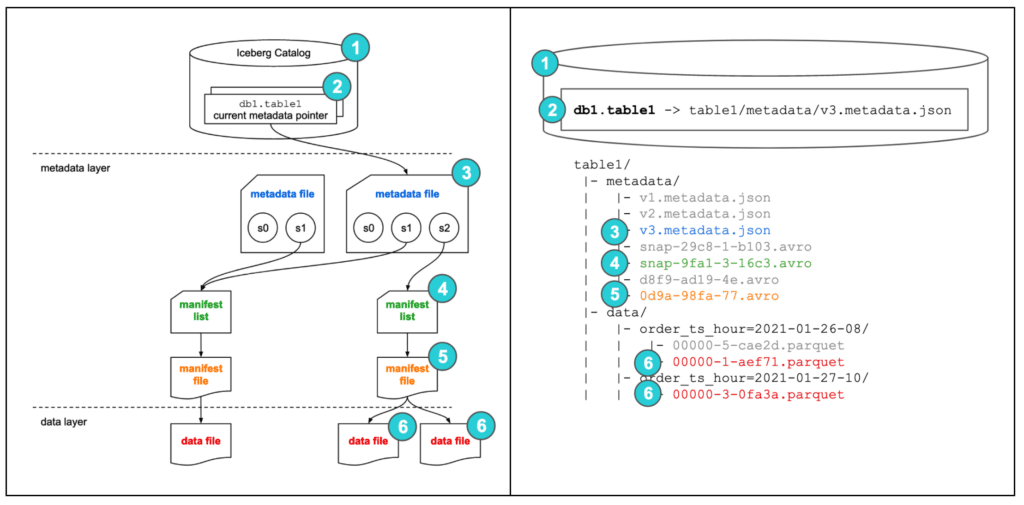
Apache Iceberg should not be seen as a storage mechanism or database. Instead it's a metadata management layer that sits on top of the data files, which we can then query later on through a dedicated and optimized query engine.
Apache Spark
Finally, a query engine... now there are MANY out there, and it's changing all the time. What also became apparent is that many of them use the commercial open-source trick, drawing you in and eventually asking you to pay for performance (which is not always a bad thing), making the choice difficult.
Seeing the customer I am working for was using Apache Spark, we thus decided to stick with it.
Personally, I think there are better alternatives popping up (hello Ray + Daft), but it currently doesn't make sense to switch and the other ecosystems still have to mature more.
Creating a CDC Implementation
Let's get started creating our actual CDC implementation. For this implementation we will thus have an orchestrator (Python) that will fetch the changes and sink them to the Iceberg maintained repository on Local Storage.
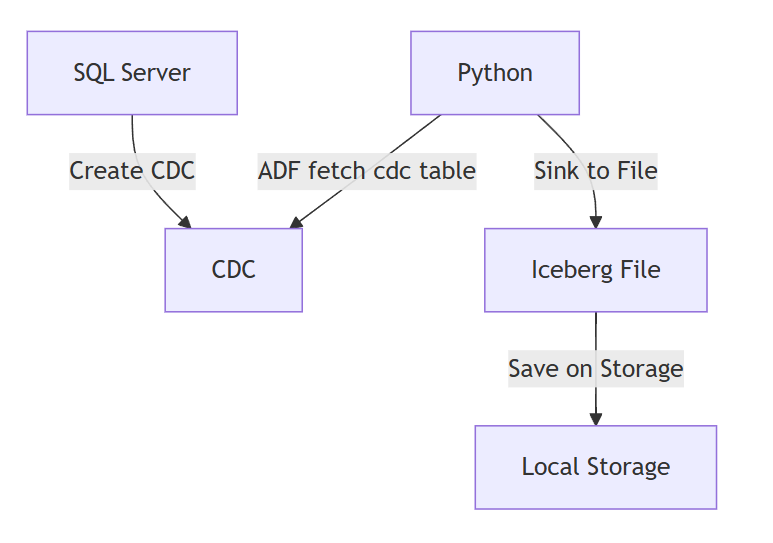
Full code: https://github.com/XavierGeerinck/PublicProjects/tree/master/Data/cdc-capture/cdc-spark
Setting up Spark
Let's create our local Spark Cluster. To do so, clone the docker-compose file from https://github.com/databricks/docker-spark-iceberg/blob/main/docker-compose.yml and spin it up:
# Download the Docker Compose file
wget https://github.com/databricks/docker-spark-iceberg/blob/main/docker-compose.yml
# Start Spark + Iceberg Cluster
docker-compose up -d
docker exec -it spark-iceberg pyspark-notebookSQL Manager
The most difficult area that I encountered is the actual SQL CDC Resource Manager, which communicate with SQL Server, connects and gets the latest changes, whereafter it merges them into Iceberg. So let's create this first.
# Fetch SQL Server CDC changes from Remote and merge them into the local Iceberg table
# we use pyodbc for this (to avoid temporary views)
import pyodbc
import sqlalchemy as sa
import polars as pl
from urllib.parse import quote_plus
from contextlib import contextmanager
LSN_DEFAULT = "0x00000000000000000000"
class SQLResource:
def __init__(self, host, db, username, password, last_lsn=None):
self.host = host
self.db = db
self.username = username
self.password = password
self.engine = sa.create_engine(
self.get_connection_string(host, db, username, password)
)
def get_connection_string(self, host, db, username, password):
"""Construct the connection string for SQLAlchemy."""
pass_escaped = quote_plus(password)
user_escaped = quote_plus(username)
driver_escaped = quote_plus("ODBC Driver 18 for SQL Server")
return f"mssql+pyodbc://{user_escaped}:{pass_escaped}@{host}/{db}?driver={driver_escaped}"
@contextmanager
def get_connection(self):
"""Get a database connection using context manager for automatic cleanup."""
connection = self.engine.connect()
try:
yield connection
finally:
connection.close()
def get_primary_key_columns(self, table_name: str) -> list[str]:
"""Get the primary key columns for a CDC-enabled table."""
with self.get_connection() as connection:
instance = self.get_capture_instance_name("dbo", table_name)
query = sa.text("""
SELECT column_name FROM cdc.index_columns WHERE object_id = (
SELECT object_id FROM cdc.change_tables WHERE capture_instance = :capture_instance_name
)
""")
result = connection.execute(query, {"capture_instance_name": instance})
primary_key_columns = [row[0] for row in result]
return primary_key_columns
def is_cdc_enabled_for_database(self):
"""Check if CDC is enabled for the database."""
with self.get_connection() as connection:
query = sa.text("""
SELECT is_cdc_enabled
FROM sys.databases
WHERE name = :db_name
""")
result = connection.execute(
query, {"db_name": self.db}
).scalar()
return bool(result)
def is_cdc_enabled_for_table(self, schema_name, table_name):
"""Check if CDC is enabled for the specified table."""
with self.get_connection() as connection:
capture_instance_name = self.get_capture_instance_name(
schema_name, table_name
)
query = sa.text(f"""
SELECT 1
FROM cdc.change_tables
WHERE capture_instance = '{capture_instance_name}'
""")
result = connection.execute(
query, {"schema_name": schema_name, "table_name": table_name}
).scalar()
return bool(result)
def get_capture_instance_name(self, schema_name, table_name):
"""Get the CDC capture instance name for a table."""
return f"dbo_{table_name}"
def get_current_lsn(self):
"""Get the current LSN from SQL Server using native function."""
with self.get_connection() as connection:
query = sa.text("SELECT sys.fn_cdc_get_max_lsn()")
return connection.execute(query).scalar()
def get_min_lsn(self, capture_instance=None):
"""Get the minimum available LSN for a capture instance."""
with self.get_connection() as connection:
query = sa.text("SELECT sys.fn_cdc_get_min_lsn(:capture_instance)")
return connection.execute(
query, {"capture_instance": capture_instance}
).scalar()
def hex_string_to_lsn(self, lsn_hex):
"""Convert a hexadecimal LSN string to binary for SQL Server functions."""
with self.get_connection() as connection:
if not lsn_hex or not isinstance(lsn_hex, str):
# Return minimum LSN if input is invalid
query = sa.text("SELECT sys.fn_cdc_get_min_lsn(NULL)")
return connection.execute(query).scalar()
if not lsn_hex.startswith("0x"):
lsn_hex = f"0x{lsn_hex}"
query = sa.text("SELECT CAST(:lsn_hex AS BINARY(10))")
result = connection.execute(query, {"lsn_hex": lsn_hex}).scalar()
if result is None:
query = sa.text("SELECT sys.fn_cdc_get_min_lsn(NULL)")
return connection.execute(query).scalar()
return result
def lsn_to_hex_string(self, lsn_bytes):
"""Convert a binary LSN to a hex string format."""
if lsn_bytes is None:
return LSN_DEFAULT
return f"0x{lsn_bytes.hex().upper()}"
def get_primary_key_columns(self, table_name: str) -> list[str]:
"""Get the primary key columns for a CDC-enabled table."""
with self.get_connection() as connection:
instance = self.get_capture_instance_name("dbo", table_name)
query = sa.text("""
SELECT column_name FROM cdc.index_columns WHERE object_id = (
SELECT object_id FROM cdc.change_tables WHERE capture_instance = :capture_instance_name
)
""")
result = connection.execute(query, {"capture_instance_name": instance})
primary_key_columns = [row[0] for row in result]
return primary_key_columns
def get_merge_predicate(self, table_name: str) -> str:
"""Uses the primary key columns to construct a predicate for merging.
e.g., CustomerID and Email become: source.CustomerID = target.CustomerID AND source.Email = target.Email
"""
primary_key_columns = self.get_primary_key_columns(table_name)
if not primary_key_columns:
raise ValueError(f"No primary key columns found for table {table_name}")
# Construct the merge predicate
merge_predicate = " AND ".join(
[f"s.{col} = t.{col}" for col in primary_key_columns]
)
return merge_predicate
def get_table_changes(
self, table_name, last_lsn=None, schema_name="dbo", chunksize=10000
) -> tuple[pl.DataFrame, str]:
"""Get changes from a CDC-enabled table since the last LSN.
Uses the native SQL Server CDC function fn_cdc_get_all_changes.
Args:
table_name (str): The name of the table to query.
last_lsn (str, optional): The last processed LSN. If None, a full copy is performed.
schema_name (str, optional): The schema name of the table. Defaults to 'dbo'.
chunksize (int, optional): Number of rows to fetch per query. Defaults to 10000.
Returns:
tuple: A tuple containing the DataFrame of changes and the current LSN.
"""
try:
with self.get_connection() as connection:
# Check if CDC is enabled for the database and table
if not self.is_cdc_enabled_for_database():
raise ValueError(
f"CDC is not enabled for database {self.config.database.get_value()}"
)
if not self.is_cdc_enabled_for_table(schema_name, table_name):
raise ValueError(
f"CDC not enabled for table {schema_name}.{table_name}"
)
# Get the capture instance name
capture_instance = self.get_capture_instance_name(
schema_name, table_name
)
if not capture_instance:
raise ValueError(
f"Could not find CDC capture instance for {schema_name}.{table_name}"
)
# Get current maximum LSN
current_lsn = self.get_current_lsn()
current_lsn_hex = self.lsn_to_hex_string(current_lsn)
# If no last_lsn provided, we should first take a first copy of the table
if last_lsn is None or last_lsn == LSN_DEFAULT:
raise ValueError(
f"Initial copy required for table {schema_name}.{table_name}"
)
# Convert LSN hex strings to binary
from_lsn_hex = last_lsn
to_lsn_hex = f"0x{current_lsn.hex()}"
# Use the native CDC function with parameterized query
# Process in chunks to avoid memory issues with large tables
query = sa.text(f"""
DECLARE @from_lsn BINARY(10), @to_lsn BINARY(10)
SET @from_lsn = CONVERT(BINARY(10), :from_lsn, 1)
SET @to_lsn = CONVERT(BINARY(10), :to_lsn, 1)
SELECT * FROM cdc.fn_cdc_get_all_changes_{capture_instance}(
@from_lsn, @to_lsn, 'all'
)
""")
# Use chunksize to process large result sets in batches
changes_df = pl.read_database(
query,
connection,
execute_options={
"parameters": {"from_lsn": from_lsn_hex, "to_lsn": to_lsn_hex}
},
)
# Convert binary LSN to hex string for storage
current_lsn_hex = self.lsn_to_hex_string(current_lsn)
return changes_df, current_lsn_hex
except Exception as e:
raise RuntimeError(
f"Database error when getting CDC changes: {str(e)}"
) from eConnecting to SQL and Pulling Changes
Once we have the manager, let's now use it and get our changes:
# Connect to SQL Server
sql_resource = SQLResource(
SQL_HOST,
SQL_DATABASE,
SQL_USER,
SQL_PASS
)
last_lsn = "0x0000004400000D280005"
changes = sql_resource.get_table_changes(
table_name=TABLE_NAME_REMOTE,
last_lsn=last_lsn, # todo: fetch this each time and save into metadata
schema_name="dbo"
)
# Make them available as temporary view
print(type(changes[0]))
changes_df = spark.createDataFrame(changes[0].to_pandas())
changes_df.createOrReplaceTempView("changes")Verifying
Verifying everything we can do by just getting the changes
print(spark.sql("SELECT * FROM changes").show())
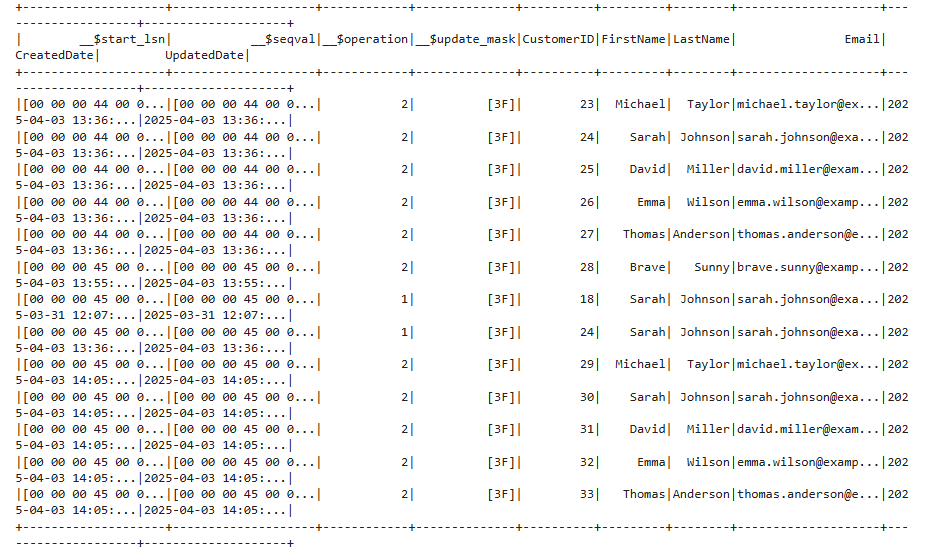
Merging the Changes
Which we merge into our Iceberg table:
# Merge the CDC changes into the iceberg table as merge
# we work with the __$operation column to determine the type of operation, which can have values:
# - Deleted (__$operation = 1),
# - Inserted (__$operation = 2)
# - Updated Before (__$operation = 3)
# - Updated After (__$operation = 4)
# https://iceberg.apache.org/docs/1.5.0/spark-writes/#merge-into
print(f"Performing merge operation on '{TABLE_NAME_LOCAL}' with predicate '{merge_predicate}'...")
spark.sql(f"""
MERGE INTO {TABLE_NAME_LOCAL} AS t
USING (SELECT * FROM changes) AS s
ON {merge_predicate}
WHEN MATCHED AND s.`__$operation` = 1 THEN DELETE
WHEN MATCHED AND s.`__$operation` IN (2, 4) THEN UPDATE SET *
-- Anything we can't match, we insert
WHEN NOT MATCHED THEN INSERT *
""")Validating
Let's run the same query and compare results + sort. We will now see that the records have changed as expected and old records have been removed, others updated. Also note that we are not merging in the __$ columns from the CDC!
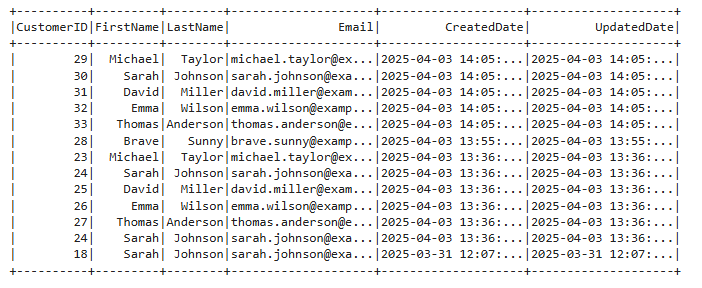
Conclusion
All the above might seem complex, but in production most of this is abstracted away for us. The most difficult part is actually integrating the CDC SQL Manager into an orchestrator (e.g., Azure Data Factory) to pull our changes in batch (or switch over to a more streaming approach if we want to work real-time). Which we can then sync through our Big Data Query Engine such as Spark into Flat Files.
Finally, what remains is to now process this flat file towards a BI application, creating a ready-to-be-consumed data warehouse.
Reference
Here you can find some amazing References that I used to come to the conclusion above.




Comments ()